The goal of bench is to benchmark code, tracking execution time, memory allocations and garbage collections.
Installation
You can install the release version from CRAN with:
install.packages("bench")Or you can install the development version from GitHub with:
# install.packages("remotes")
remotes::install_github("r-lib/bench")Features
bench::mark() is used to benchmark one or a series of expressions, we feel it has a number of advantages over alternatives.
- Always uses the highest precision APIs available for each operating system (often nanoseconds).
- Tracks memory allocations for each expression.
- Tracks the number and type of R garbage collections per expression iteration.
- Verifies equality of expression results by default, to avoid accidentally benchmarking inequivalent code.
- Has
bench::press(), which allows you to easily perform and combine benchmarks across a large grid of values. - Uses adaptive stopping by default, running each expression for a set amount of time rather than for a specific number of iterations.
- Expressions are run in batches and summary statistics are calculated after filtering out iterations with garbage collections. This allows you to isolate the performance and effects of garbage collection on running time (for more details see Neal 2014).
The times and memory usage are returned as custom objects which have human readable formatting for display (e.g. 104ns) and comparisons (e.g. x$mem_alloc > "10MB").
There is also full support for plotting with ggplot2 including custom scales and formatting.
Usage
bench::mark()
Benchmarks can be run with bench::mark(), which takes one or more expressions to benchmark against each other.
bench::mark() will throw an error if the results are not equivalent, so you don’t accidentally benchmark inequivalent code.
bench::mark(
dat[dat$x > 500, ],
dat[which(dat$x > 499), ],
subset(dat, x > 500))
#> Error: Each result must equal the first result:
#> `dat[dat$x > 500, ]` does not equal `dat[which(dat$x > 499), ]`Results are easy to interpret, with human readable units.
bnch <- bench::mark(
dat[dat$x > 500, ],
dat[which(dat$x > 500), ],
subset(dat, x > 500))
bnch
#> # A tibble: 3 × 6
#> expression min median `itr/sec` mem_alloc `gc/sec`
#> <bch:expr> <bch:tm> <bch:tm> <dbl> <bch:byt> <dbl>
#> 1 dat[dat$x > 500, ] 309µs 423µs 2392. 377KB 15.0
#> 2 dat[which(dat$x > 500), ] 212µs 279µs 3406. 260KB 14.8
#> 3 subset(dat, x > 500) 380µs 446µs 2141. 510KB 17.9By default the summary uses absolute measures, however relative results can be obtained by using relative = TRUE in your call to bench::mark() or calling summary(relative = TRUE) on the results.
summary(bnch, relative = TRUE)
#> # A tibble: 3 × 6
#> expression min median `itr/sec` mem_alloc `gc/sec`
#> <bch:expr> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 dat[dat$x > 500, ] 1.46 1.52 1.12 1.45 1.01
#> 2 dat[which(dat$x > 500), ] 1 1 1.59 1 1
#> 3 subset(dat, x > 500) 1.79 1.60 1 1.96 1.21
bench::press()
bench::press() is used to run benchmarks against a grid of parameters. Provide setup and benchmarking code as a single unnamed argument then define sets of values as named arguments. The full combination of values will be expanded and the benchmarks are then pressed together in the result. This allows you to benchmark a set of expressions across a wide variety of input sizes, perform replications and other useful tasks.
set.seed(42)
create_df <- function(rows, cols) {
as.data.frame(setNames(
replicate(cols, runif(rows, 1, 100), simplify = FALSE),
rep_len(c("x", letters), cols)))
}
results <- bench::press(
rows = c(1000, 10000),
cols = c(2, 10),
{
dat <- create_df(rows, cols)
bench::mark(
min_iterations = 100,
bracket = dat[dat$x > 500, ],
which = dat[which(dat$x > 500), ],
subset = subset(dat, x > 500)
)
}
)
#> Running with:
#> rows cols
#> 1 1000 2
#> 2 10000 2
#> 3 1000 10
#> 4 10000 10
results
#> # A tibble: 12 × 8
#> expression rows cols min median `itr/sec` mem_alloc `gc/sec`
#> <bch:expr> <dbl> <dbl> <bch:tm> <bch:tm> <dbl> <bch:byt> <dbl>
#> 1 bracket 1000 2 25.3µs 33µs 29008. 15.84KB 11.6
#> 2 which 1000 2 25.5µs 32.5µs 29813. 7.91KB 8.95
#> 3 subset 1000 2 45.8µs 56.7µs 17164. 27.7KB 8.66
#> 4 bracket 10000 2 45.6µs 60µs 16286. 156.46KB 56.1
#> 5 which 10000 2 40.4µs 42.9µs 20583. 78.23KB 31.7
#> 6 subset 10000 2 94.6µs 117.5µs 8158. 273.79KB 49.1
#> 7 bracket 1000 10 64.4µs 77µs 12601. 47.52KB 12.8
#> 8 which 1000 10 58.9µs 63.3µs 14338. 7.91KB 12.4
#> 9 subset 1000 10 85.1µs 104.7µs 8755. 59.38KB 10.7
#> 10 bracket 10000 10 128.9µs 144.6µs 6752. 469.4KB 70.3
#> 11 which 10000 10 89.8µs 97.3µs 9699. 78.23KB 14.8
#> 12 subset 10000 10 189.5µs 232.9µs 4180. 586.73KB 56.8Plotting
ggplot2::autoplot() can be used to generate an informative default plot. This plot is colored by gc level (0, 1, or 2) and faceted by parameters (if any). By default it generates a beeswarm plot, however you can also specify other plot types (jitter, ridge, boxplot, violin). See ?autoplot.bench_mark for full details.
ggplot2::autoplot(results)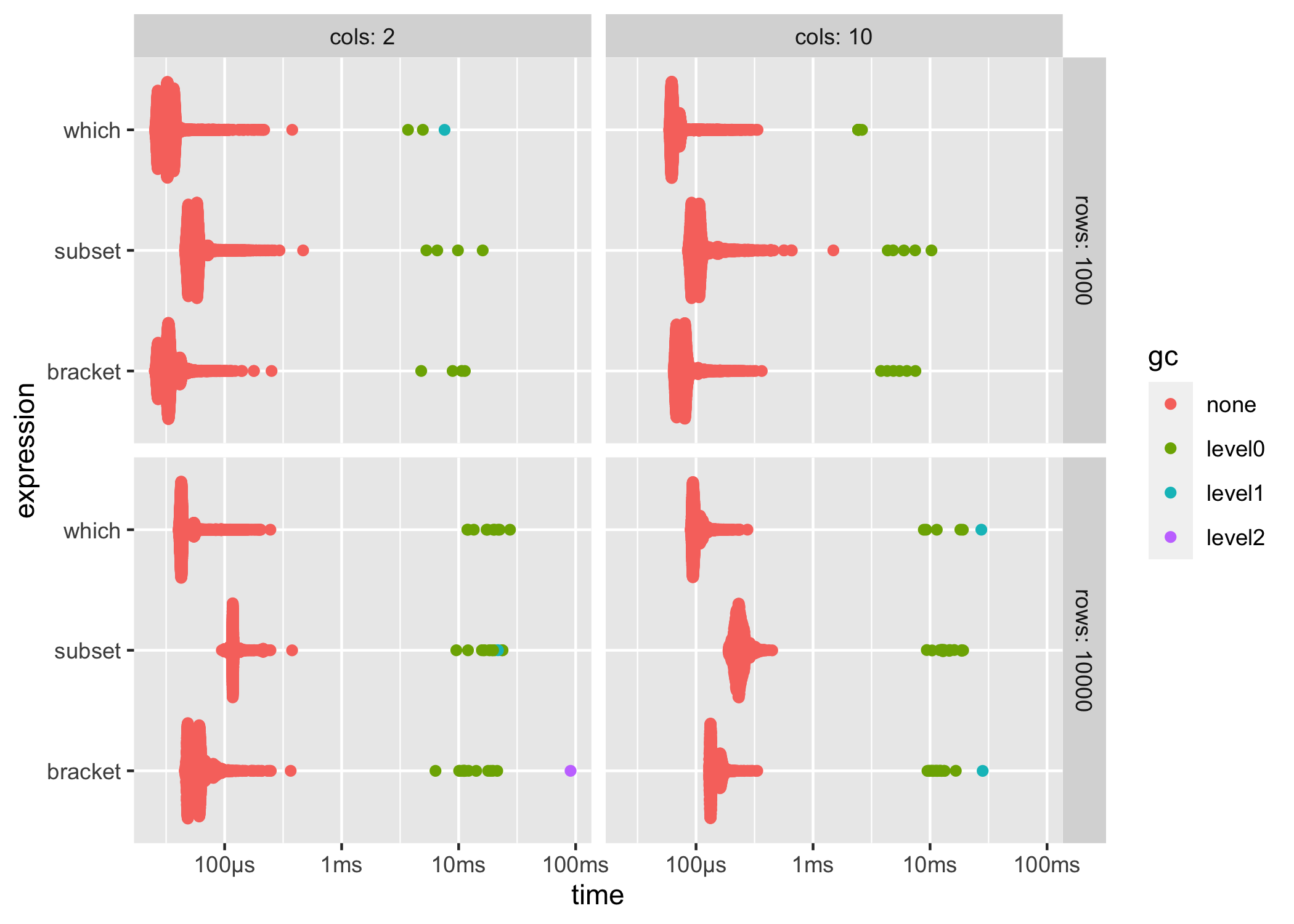
You can also produce fully custom plots by un-nesting the results and working with the data directly.
system_time()
bench also includes system_time(), a higher precision alternative to system.time().
bench::system_time({ i <- 1; while(i < 1e7) i <- i + 1 })
#> process real
#> 2.58s 2.59s
bench::system_time(Sys.sleep(.5))
#> process real
#> 73µs 500ms