Tidy summarizes information about the components of a model. A model component might be a single term in a regression, a single hypothesis, a cluster, or a class. Exactly what tidy considers to be a model component varies across models but is usually self-evident. If a model has several distinct types of components, you will need to specify which components to return.
# S3 method for glht tidy(x, conf.int = FALSE, conf.level = 0.95, ...)
Arguments
| x | A |
|---|---|
| conf.int | Logical indicating whether or not to include a confidence
interval in the tidied output. Defaults to |
| conf.level | The confidence level to use for the confidence interval
if |
| ... | Additional arguments. Not used. Needed to match generic
signature only. Cautionary note: Misspelled arguments will be
absorbed in |
See also
Other multcomp tidiers:
tidy.cld(),
tidy.confint.glht(),
tidy.summary.glht()
Value
A tibble::tibble() with columns:
Levels being compared.
The estimated value of the regression term.
Value to which the estimate is compared.
Examples
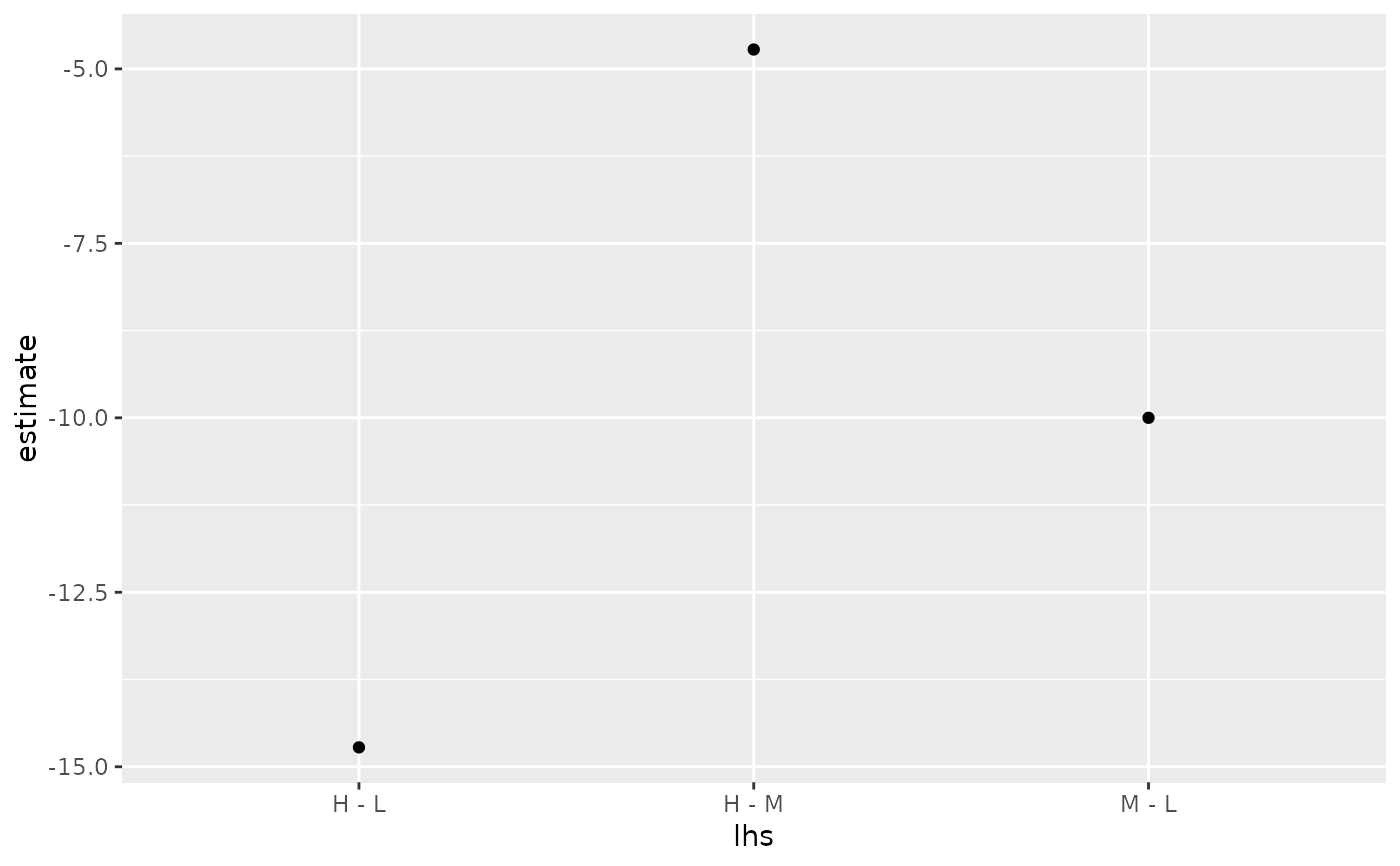
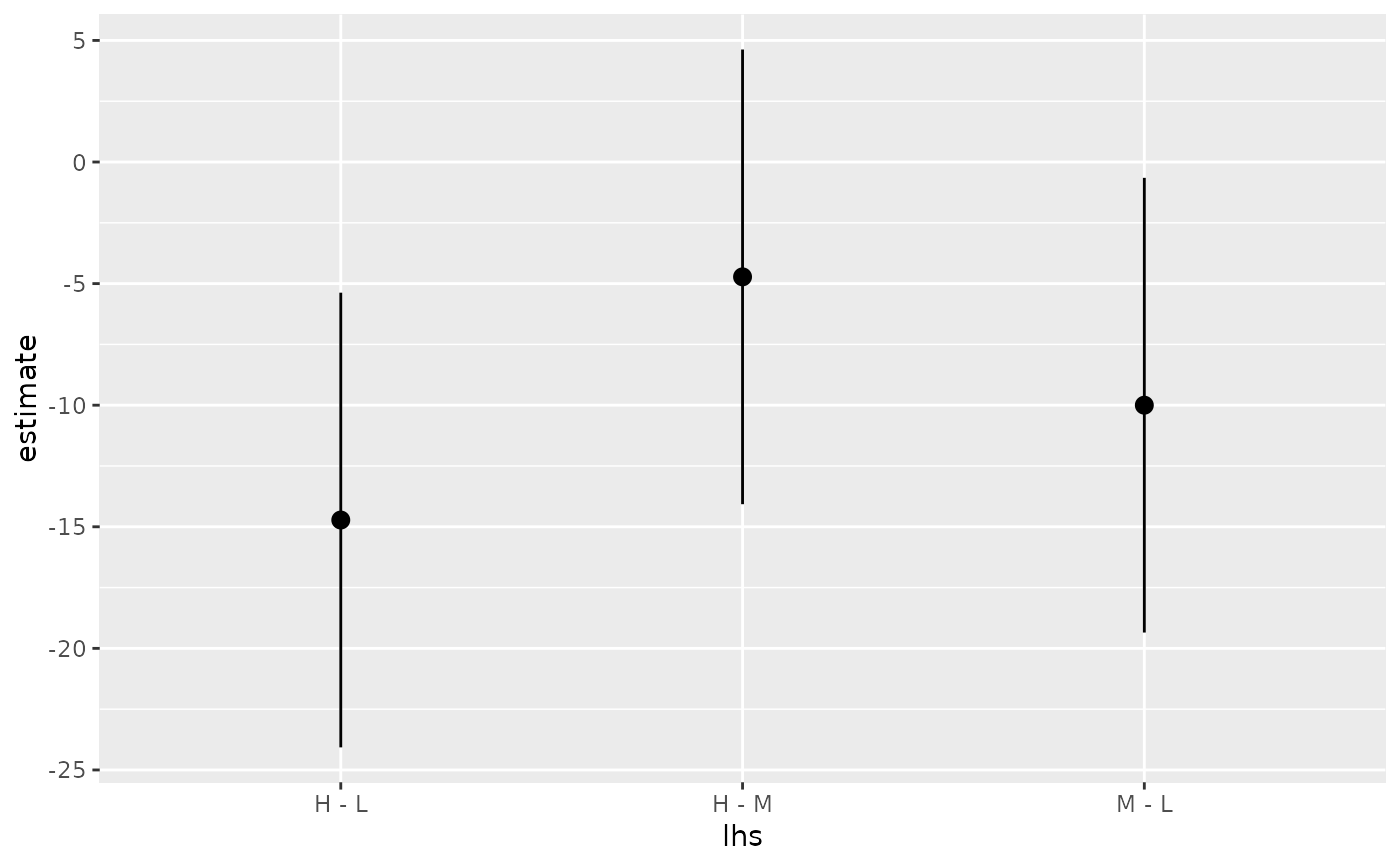
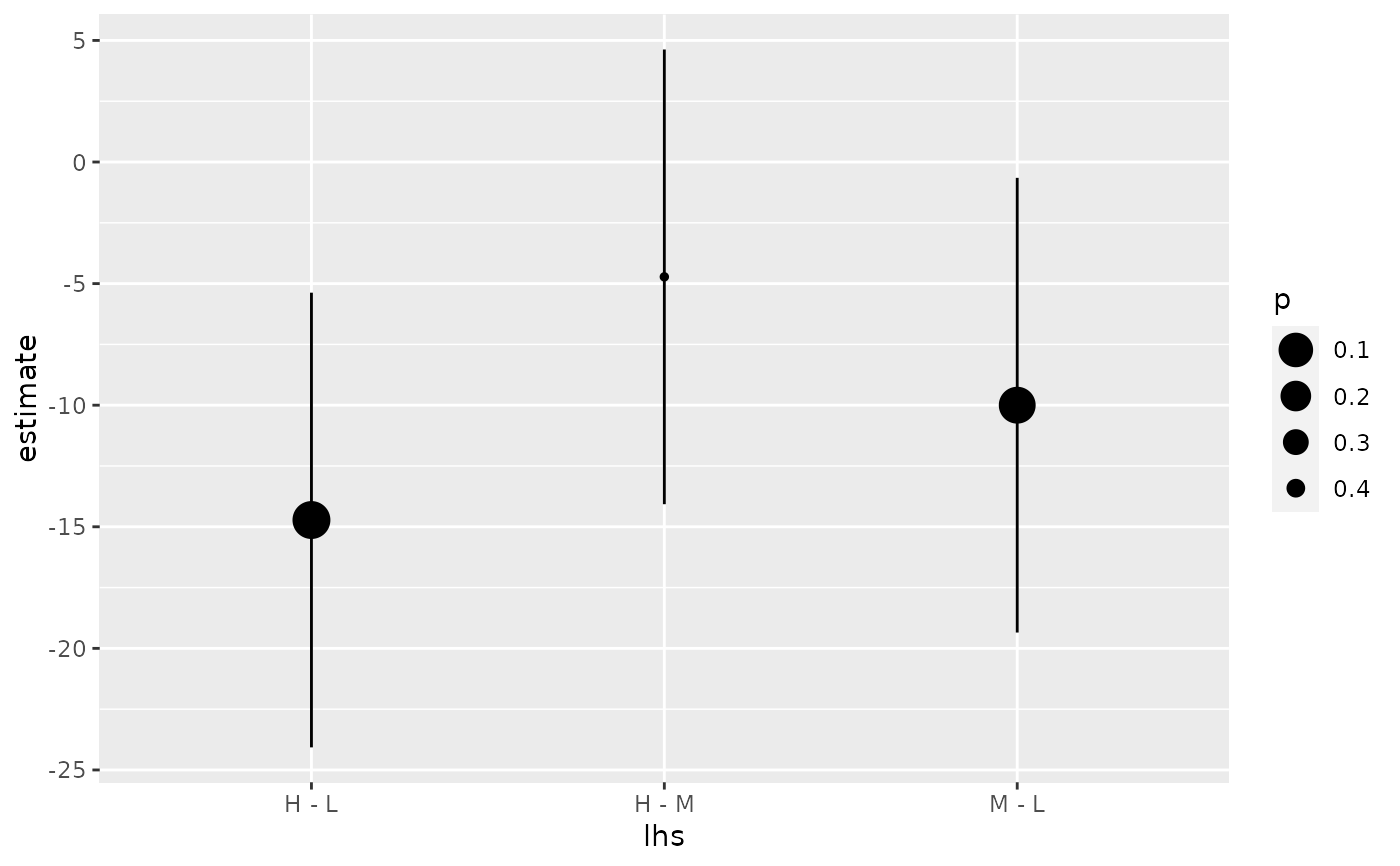
library(multcomp) library(ggplot2) amod <- aov(breaks ~ wool + tension, data = warpbreaks) wht <- glht(amod, linfct = mcp(tension = "Tukey")) tidy(wht)#> # A tibble: 3 x 7 #> term contrast null.value estimate std.error statistic adj.p.value #> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> #> 1 tension M - L 0 -10. 3.87 -2.58 0.0336 #> 2 tension H - L 0 -14.7 3.87 -3.80 0.00110 #> 3 tension H - M 0 -4.72 3.87 -1.22 0.447#> # A tibble: 3 x 5 #> term contrast estimate conf.low conf.high #> <chr> <chr> <dbl> <dbl> <dbl> #> 1 tension M - L -10. -19.3 -0.651 #> 2 tension H - L -14.7 -24.1 -5.37 #> 3 tension H - M -4.72 -14.1 4.63#> # A tibble: 3 x 7 #> term contrast null.value estimate std.error statistic adj.p.value #> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> #> 1 tension M - L 0 -10. 3.87 -2.58 0.0336 #> 2 tension H - L 0 -14.7 3.87 -3.80 0.00110 #> 3 tension H - M 0 -4.72 3.87 -1.22 0.447ggplot(mapping = aes(lhs, estimate)) + geom_linerange(aes(ymin = lwr, ymax = upr), data = CI) + geom_point(aes(size = p), data = summary(wht)) + scale_size(trans = "reverse")#> # A tibble: 3 x 2 #> tension letters #> <chr> <chr> #> 1 L b #> 2 M a #> 3 H a