Glance accepts a model object and returns a tibble::tibble()
with exactly one row of model summaries. The summaries are typically
goodness of fit measures, p-values for hypothesis tests on residuals,
or model convergence information.
Glance never returns information from the original call to the modeling function. This includes the name of the modeling function or any arguments passed to the modeling function.
Glance does not calculate summary measures. Rather, it farms out these
computations to appropriate methods and gathers the results together.
Sometimes a goodness of fit measure will be undefined. In these cases
the measure will be reported as NA.
Glance returns the same number of columns regardless of whether the
model matrix is rank-deficient or not. If so, entries in columns
that no longer have a well-defined value are filled in with an NA
of the appropriate type.
# S3 method for survfit glance(x, ...)
Arguments
| x | An |
|---|---|
| ... | Additional arguments passed to |
See also
Other cch tidiers:
glance.cch(),
tidy.cch()
Other survival tidiers:
augment.coxph(),
augment.survreg(),
glance.aareg(),
glance.cch(),
glance.coxph(),
glance.pyears(),
glance.survdiff(),
glance.survexp(),
glance.survreg(),
tidy.aareg(),
tidy.cch(),
tidy.coxph(),
tidy.pyears(),
tidy.survdiff(),
tidy.survexp(),
tidy.survfit(),
tidy.survreg()
Value
A tibble::tibble() with exactly one row and columns:
Number of events.
Maximum number of subjects at risk.
Initial number of subjects at risk.
Number of observations used.
Number of observations
Restricted mean (see [survival::print.survfit()]).
Restricted mean standard error.
lower end of confidence interval on median
upper end of confidence interval on median
median survival
Examples
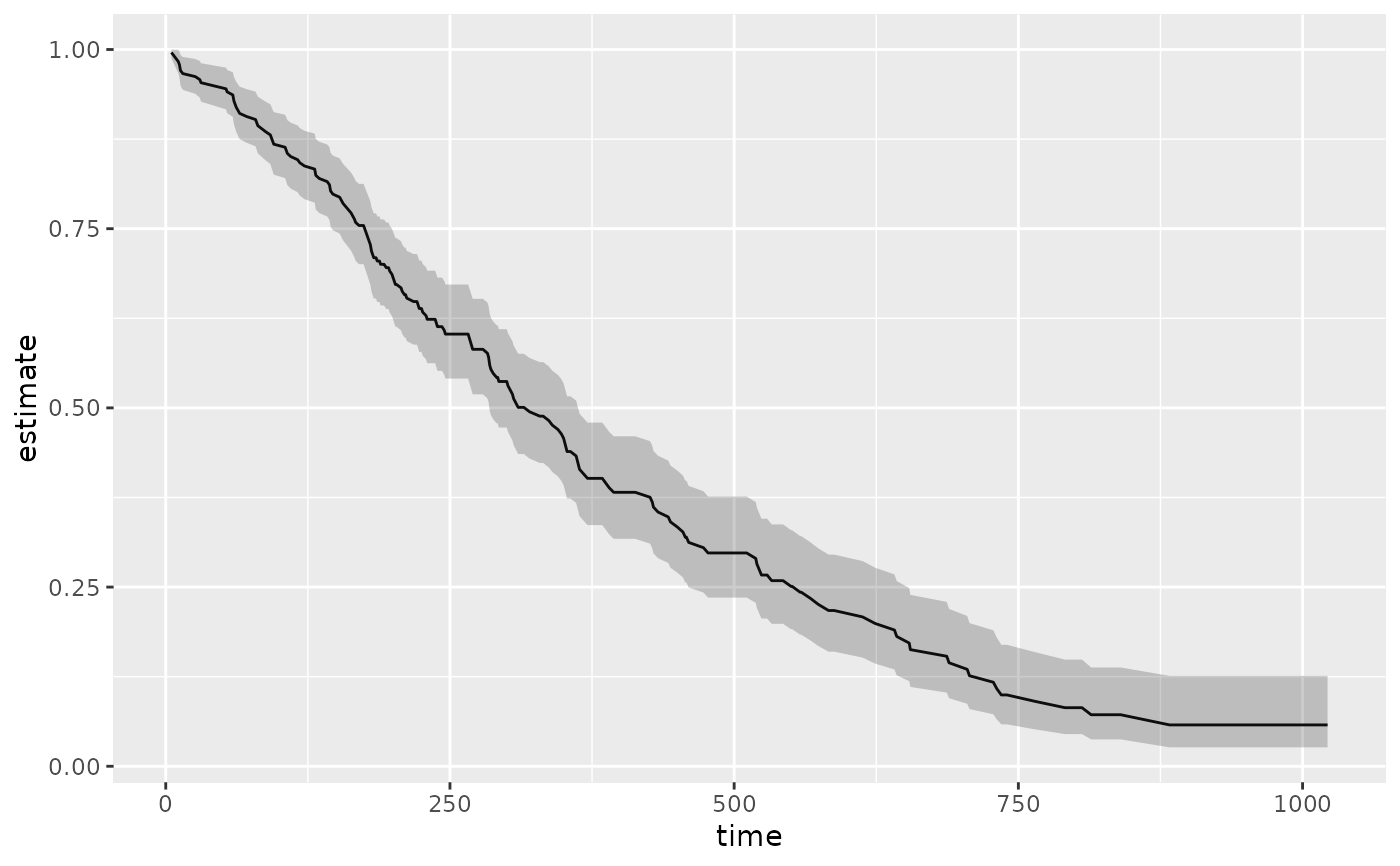
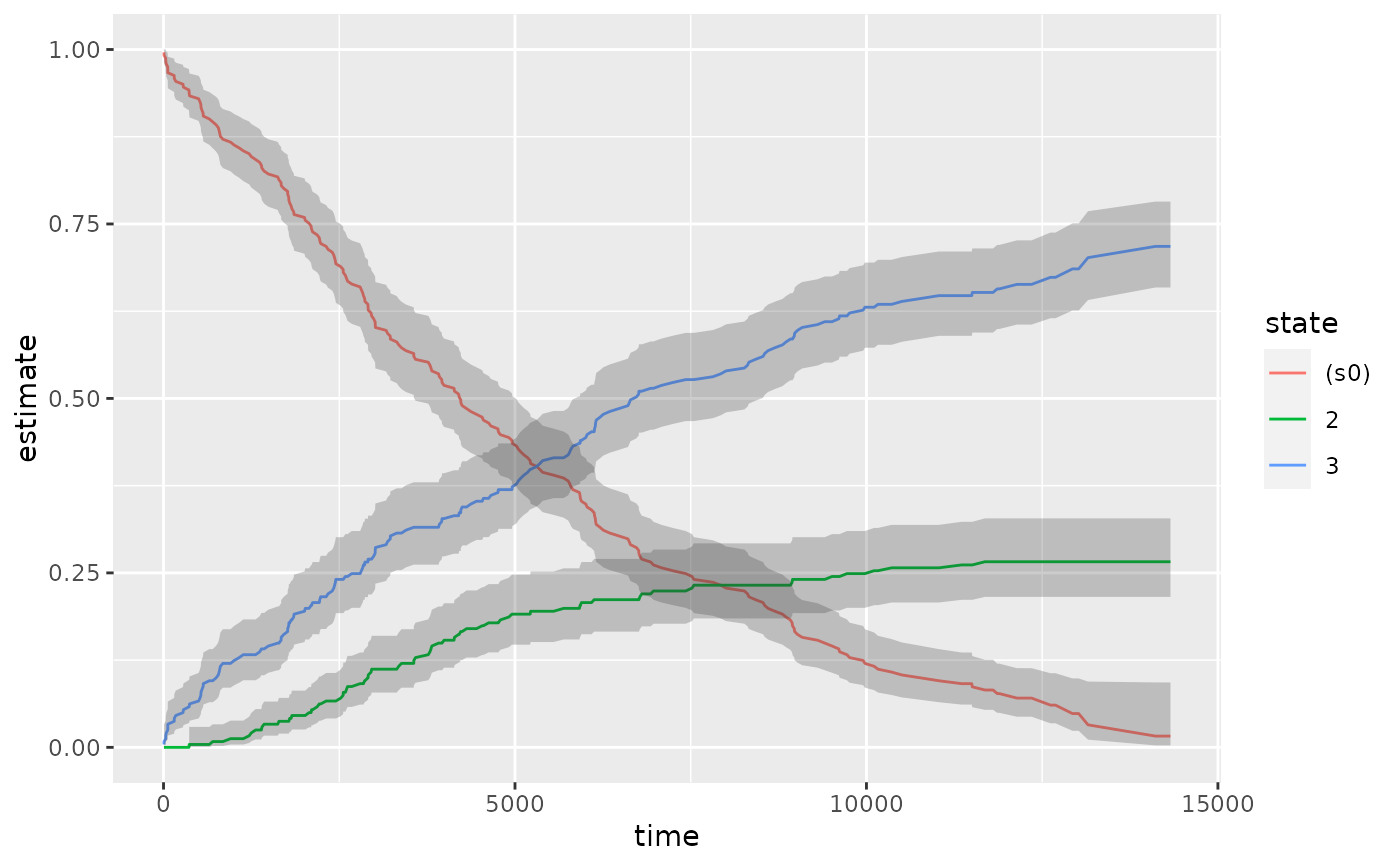
library(survival) cfit <- coxph(Surv(time, status) ~ age + sex, lung) sfit <- survfit(cfit) tidy(sfit)#> # A tibble: 186 x 8 #> time n.risk n.event n.censor estimate std.error conf.high conf.low #> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> #> 1 5 228 1 0 0.996 0.00419 1 0.988 #> 2 11 227 3 0 0.983 0.00845 1.00 0.967 #> 3 12 224 1 0 0.979 0.00947 0.997 0.961 #> 4 13 223 2 0 0.971 0.0113 0.992 0.949 #> 5 15 221 1 0 0.966 0.0121 0.990 0.944 #> 6 26 220 1 0 0.962 0.0129 0.987 0.938 #> 7 30 219 1 0 0.958 0.0136 0.984 0.933 #> 8 31 218 1 0 0.954 0.0143 0.981 0.927 #> 9 53 217 2 0 0.945 0.0157 0.975 0.917 #> 10 54 215 1 0 0.941 0.0163 0.972 0.911 #> # … with 176 more rows#> # A tibble: 1 x 10 #> records n.max n.start events rmean rmean.std.error median conf.low conf.high #> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> #> 1 228 228 228 165 381. 20.3 320 285 363 #> # … with 1 more variable: nobs <int>library(ggplot2) ggplot(tidy(sfit), aes(time, estimate)) + geom_line() + geom_ribbon(aes(ymin = conf.low, ymax = conf.high), alpha = .25)# multi-state fitCI <- survfit(Surv(stop, status * as.numeric(event), type = "mstate") ~ 1, data = mgus1, subset = (start == 0) ) td_multi <- tidy(fitCI) td_multi#> # A tibble: 711 x 9 #> time n.risk n.event n.censor estimate std.error conf.high conf.low state #> <dbl> <int> <int> <int> <dbl> <dbl> <dbl> <dbl> <fct> #> 1 6 241 0 0 0.996 0.00414 1 0.988 (s0) #> 2 7 240 0 0 0.992 0.00584 1 0.980 (s0) #> 3 31 239 0 0 0.988 0.00714 1 0.974 (s0) #> 4 32 238 0 0 0.983 0.00823 1.00 0.967 (s0) #> 5 39 237 0 0 0.979 0.00918 0.997 0.961 (s0) #> 6 60 236 0 0 0.975 0.0100 0.995 0.956 (s0) #> 7 61 235 0 0 0.967 0.0115 0.990 0.944 (s0) #> 8 152 233 0 0 0.963 0.0122 0.987 0.939 (s0) #> 9 153 232 0 0 0.959 0.0128 0.984 0.934 (s0) #> 10 174 231 0 0 0.954 0.0134 0.981 0.928 (s0) #> # … with 701 more rowsggplot(td_multi, aes(time, estimate, group = state)) + geom_line(aes(color = state)) + geom_ribbon(aes(ymin = conf.low, ymax = conf.high), alpha = .25)