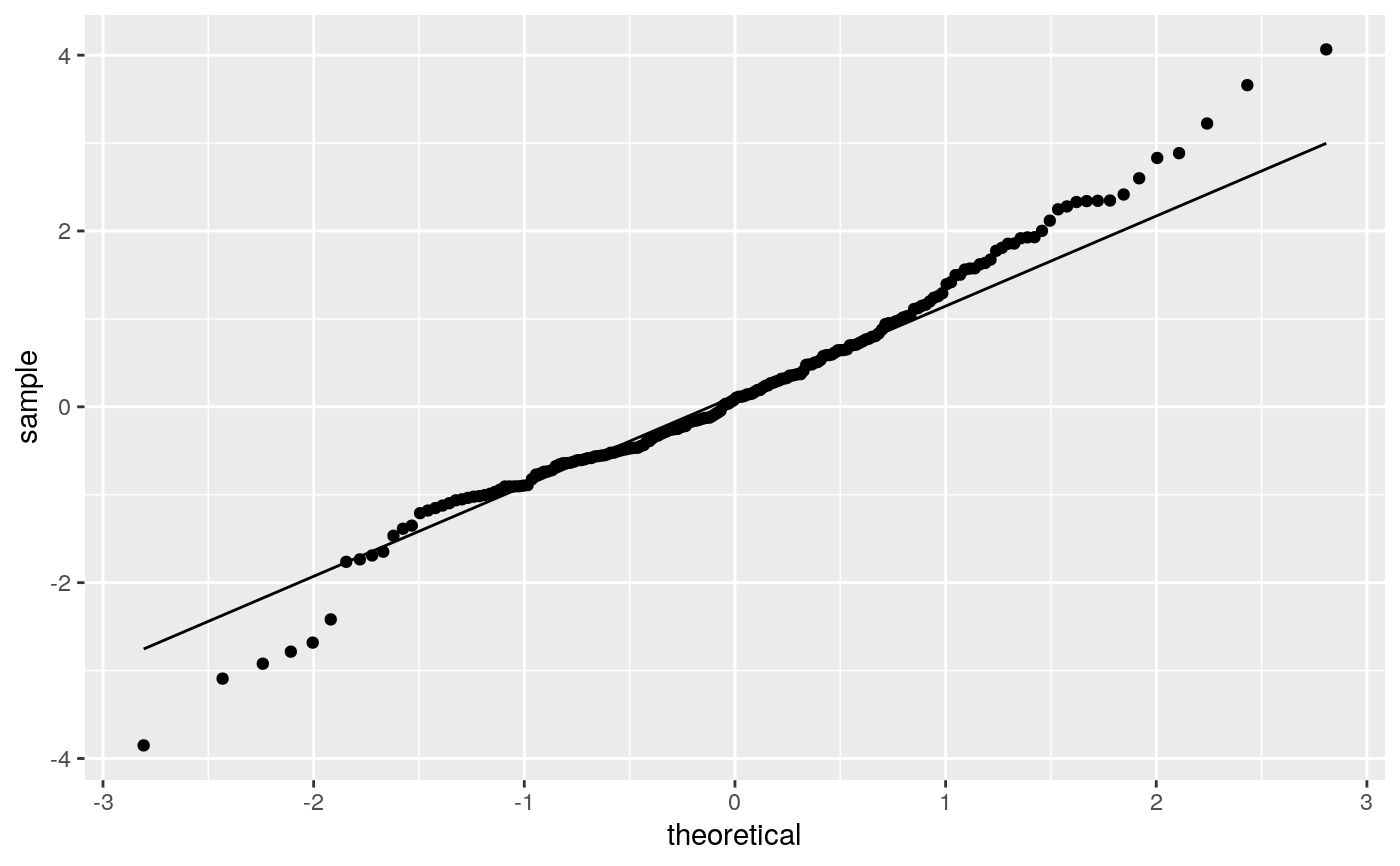
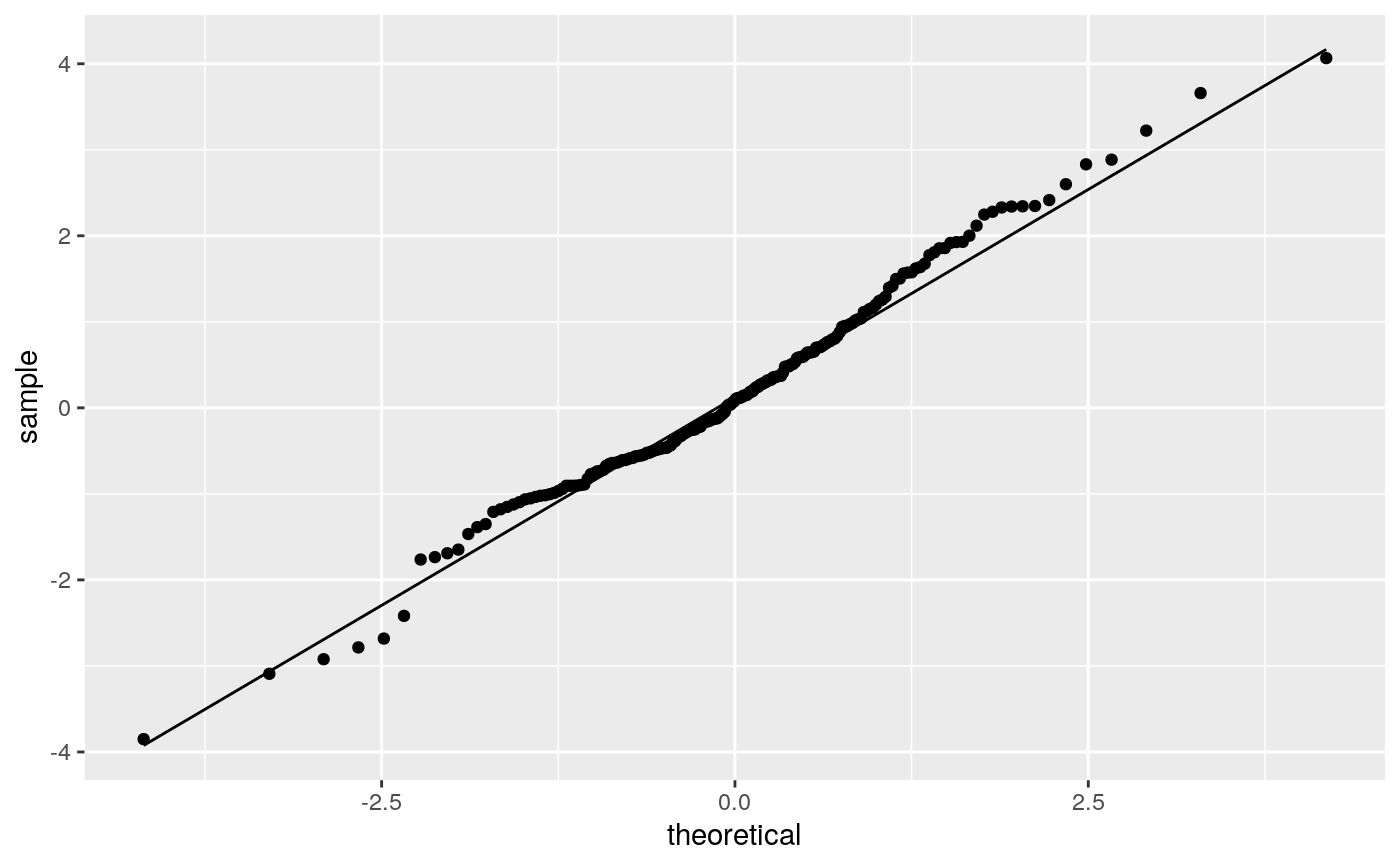
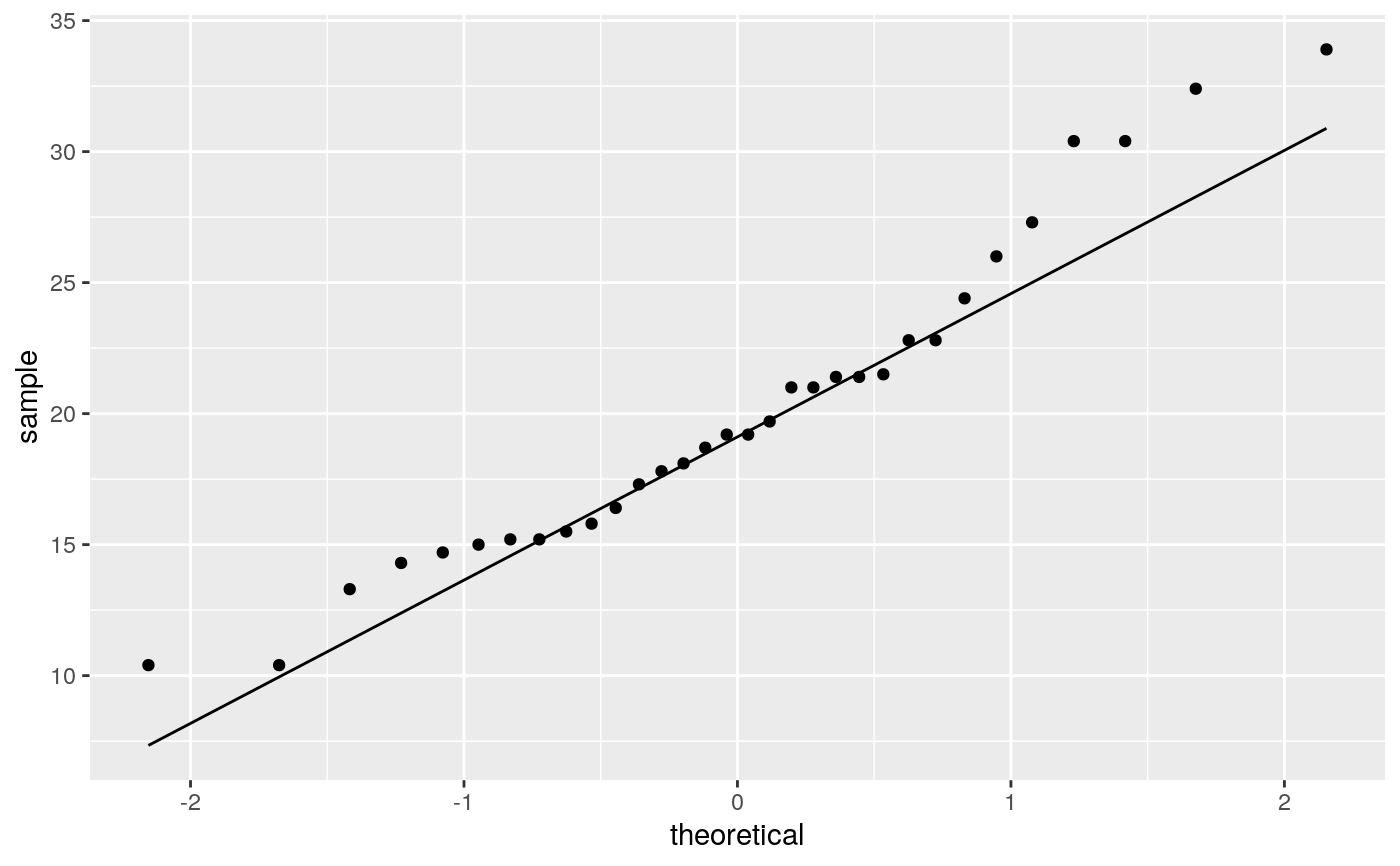
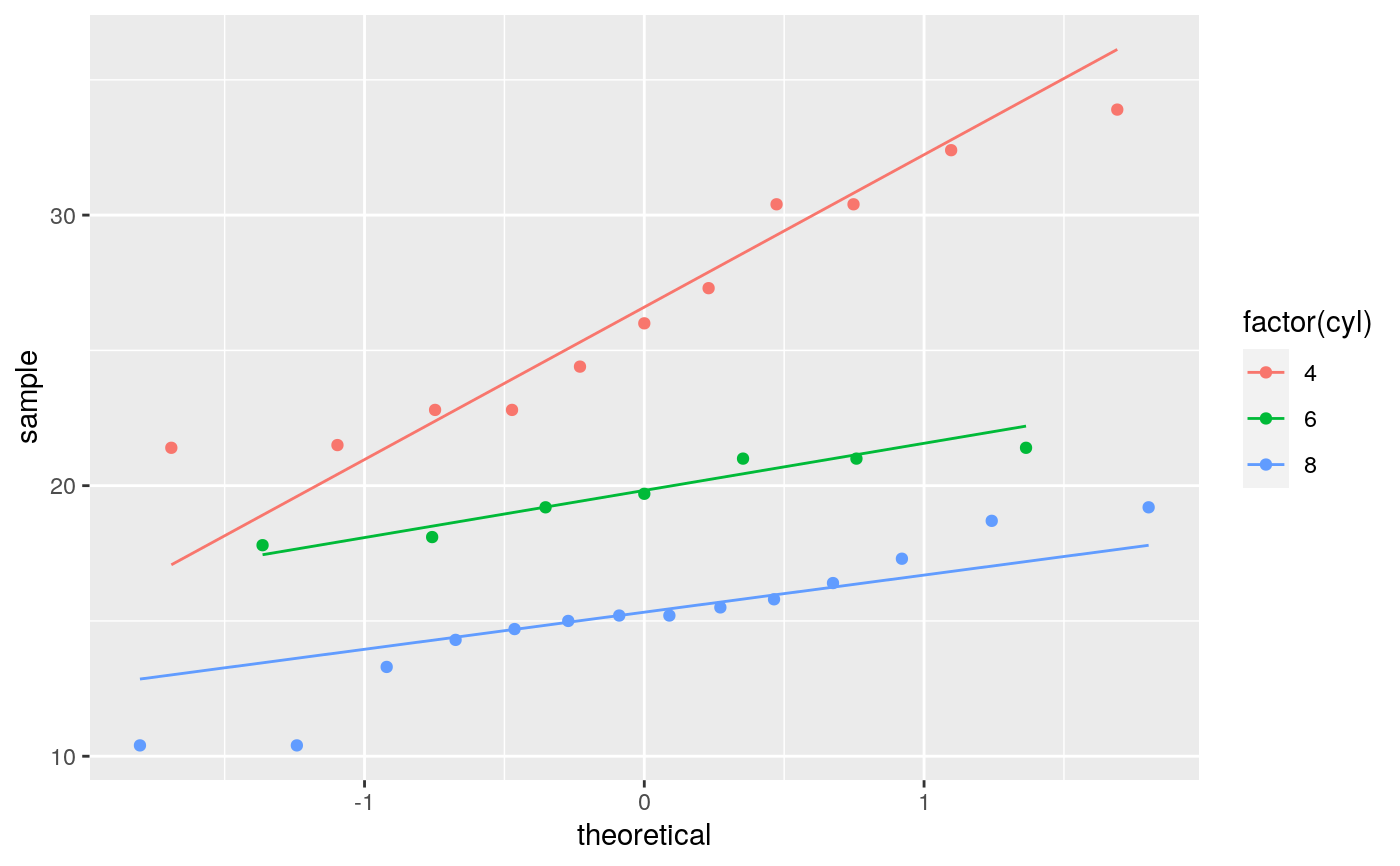
geom_qq and stat_qq produce quantile-quantile plots. geom_qq_line and
stat_qq_line compute the slope and intercept of the line connecting the
points at specified quartiles of the theoretical and sample distributions.
geom_qq_line( mapping = NULL, data = NULL, geom = "path", position = "identity", ..., distribution = stats::qnorm, dparams = list(), line.p = c(0.25, 0.75), fullrange = FALSE, na.rm = FALSE, show.legend = NA, inherit.aes = TRUE ) stat_qq_line( mapping = NULL, data = NULL, geom = "path", position = "identity", ..., distribution = stats::qnorm, dparams = list(), line.p = c(0.25, 0.75), fullrange = FALSE, na.rm = FALSE, show.legend = NA, inherit.aes = TRUE ) geom_qq( mapping = NULL, data = NULL, geom = "point", position = "identity", ..., distribution = stats::qnorm, dparams = list(), na.rm = FALSE, show.legend = NA, inherit.aes = TRUE ) stat_qq( mapping = NULL, data = NULL, geom = "point", position = "identity", ..., distribution = stats::qnorm, dparams = list(), na.rm = FALSE, show.legend = NA, inherit.aes = TRUE )
Arguments
| mapping | Set of aesthetic mappings created by |
|---|---|
| data | The data to be displayed in this layer. There are three options: If A A |
| geom | The geometric object to use display the data |
| position | Position adjustment, either as a string, or the result of a call to a position adjustment function. |
| ... | Other arguments passed on to |
| distribution | Distribution function to use, if x not specified |
| dparams | Additional parameters passed on to |
| line.p | Vector of quantiles to use when fitting the Q-Q line, defaults
defaults to |
| fullrange | Should the q-q line span the full range of the plot, or just the data |
| na.rm | If |
| show.legend | logical. Should this layer be included in the legends?
|
| inherit.aes | If |
Aesthetics
stat_qq() understands the following aesthetics (required aesthetics are in bold):
samplegroupxy
Learn more about setting these aesthetics in vignette("ggplot2-specs").
stat_qq_line() understands the following aesthetics (required aesthetics are in bold):
samplegroupxy
Learn more about setting these aesthetics in vignette("ggplot2-specs").
Computed variables
Variables computed by stat_qq:
sample quantiles
theoretical quantiles
Variables computed by stat_qq_line:
x-coordinates of the endpoints of the line segment connecting the points at the chosen quantiles of the theoretical and the sample distributions
y-coordinates of the endpoints
Examples
# \donttest{ df <- data.frame(y = rt(200, df = 5)) p <- ggplot(df, aes(sample = y)) p + stat_qq() + stat_qq_line()# Use fitdistr from MASS to estimate distribution params params <- as.list(MASS::fitdistr(df$y, "t")$estimate) ggplot(df, aes(sample = y)) + stat_qq(distribution = qt, dparams = params["df"]) + stat_qq_line(distribution = qt, dparams = params["df"])# Using to explore the distribution of a variable ggplot(mtcars, aes(sample = mpg)) + stat_qq() + stat_qq_line()# }