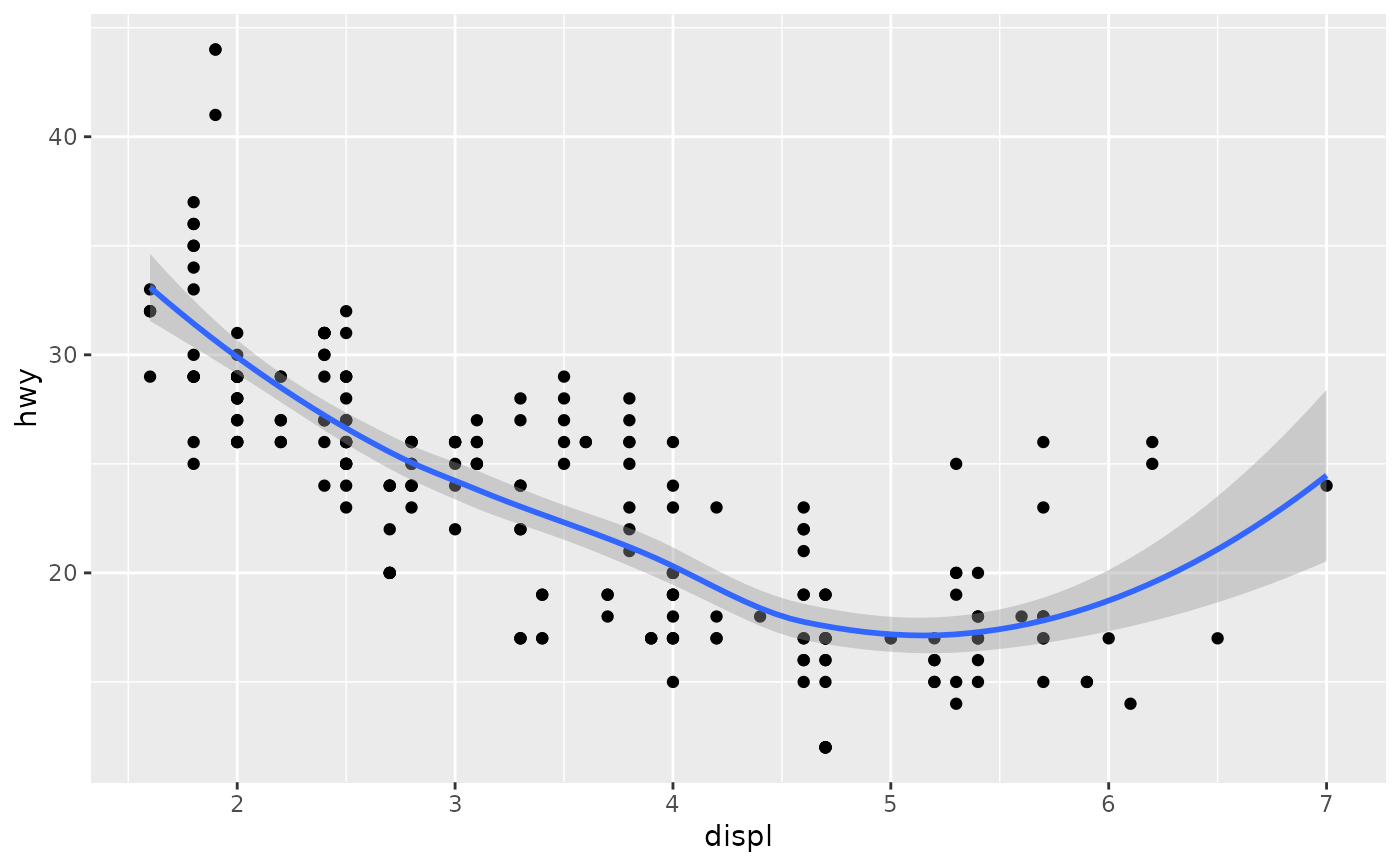
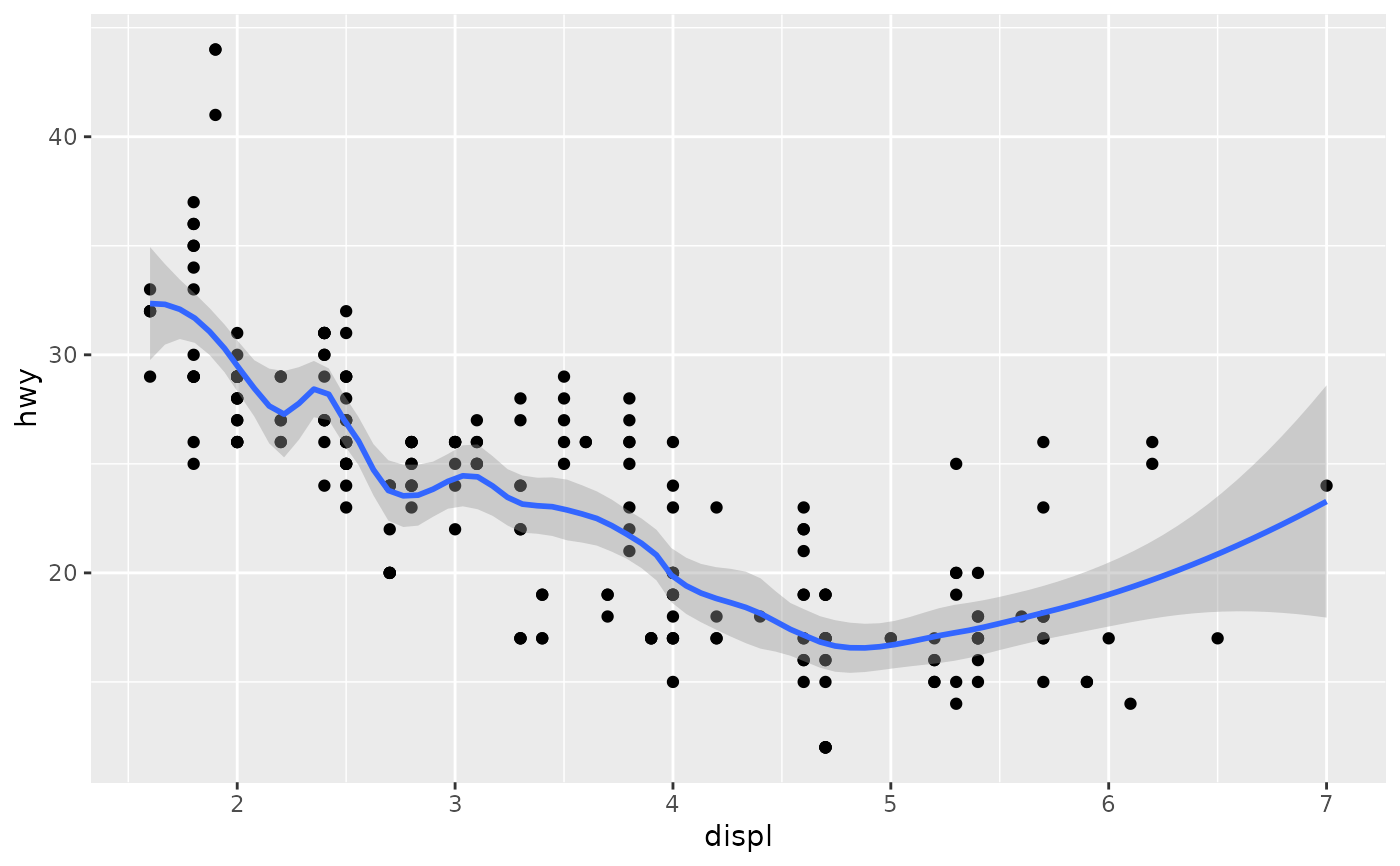
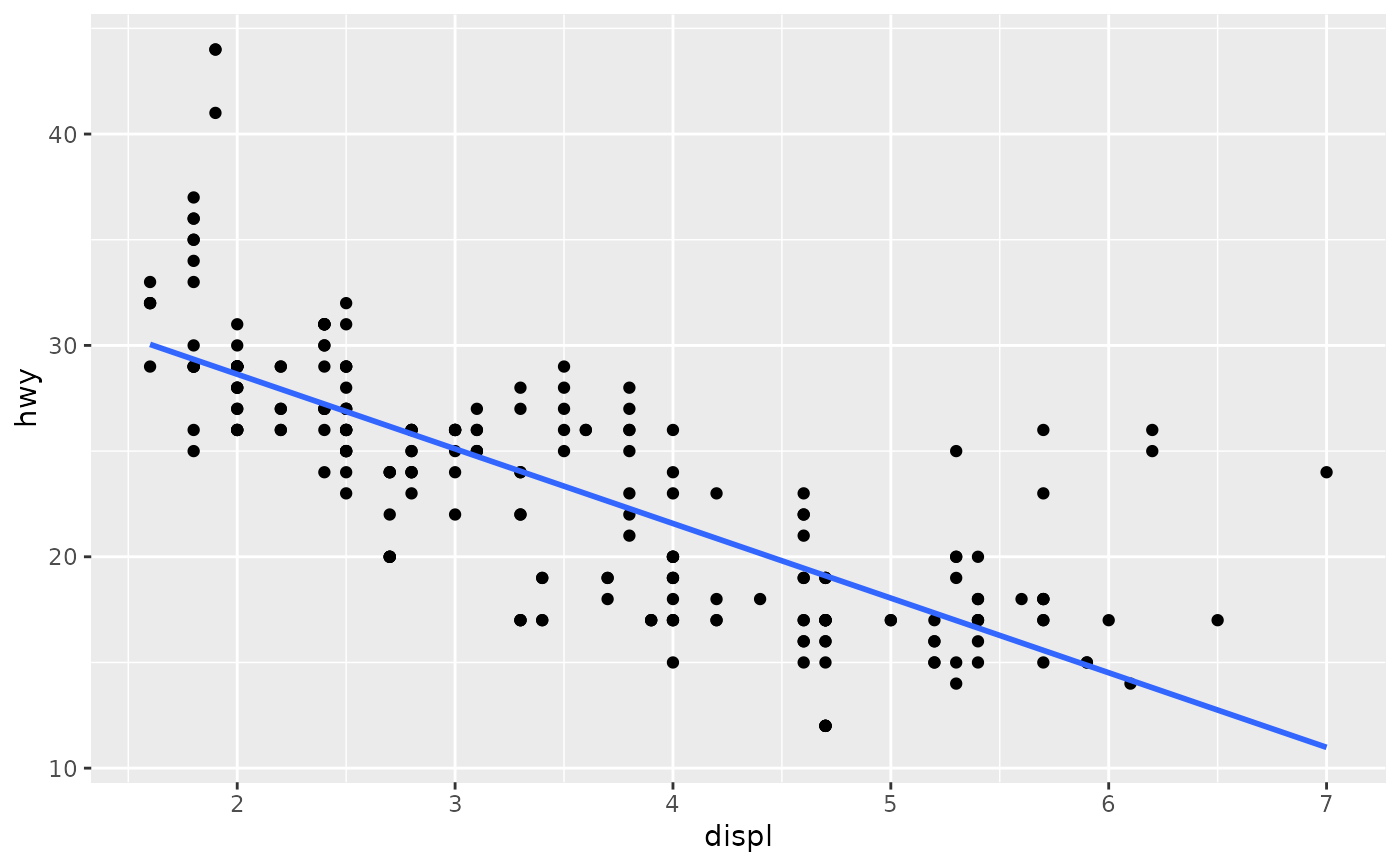
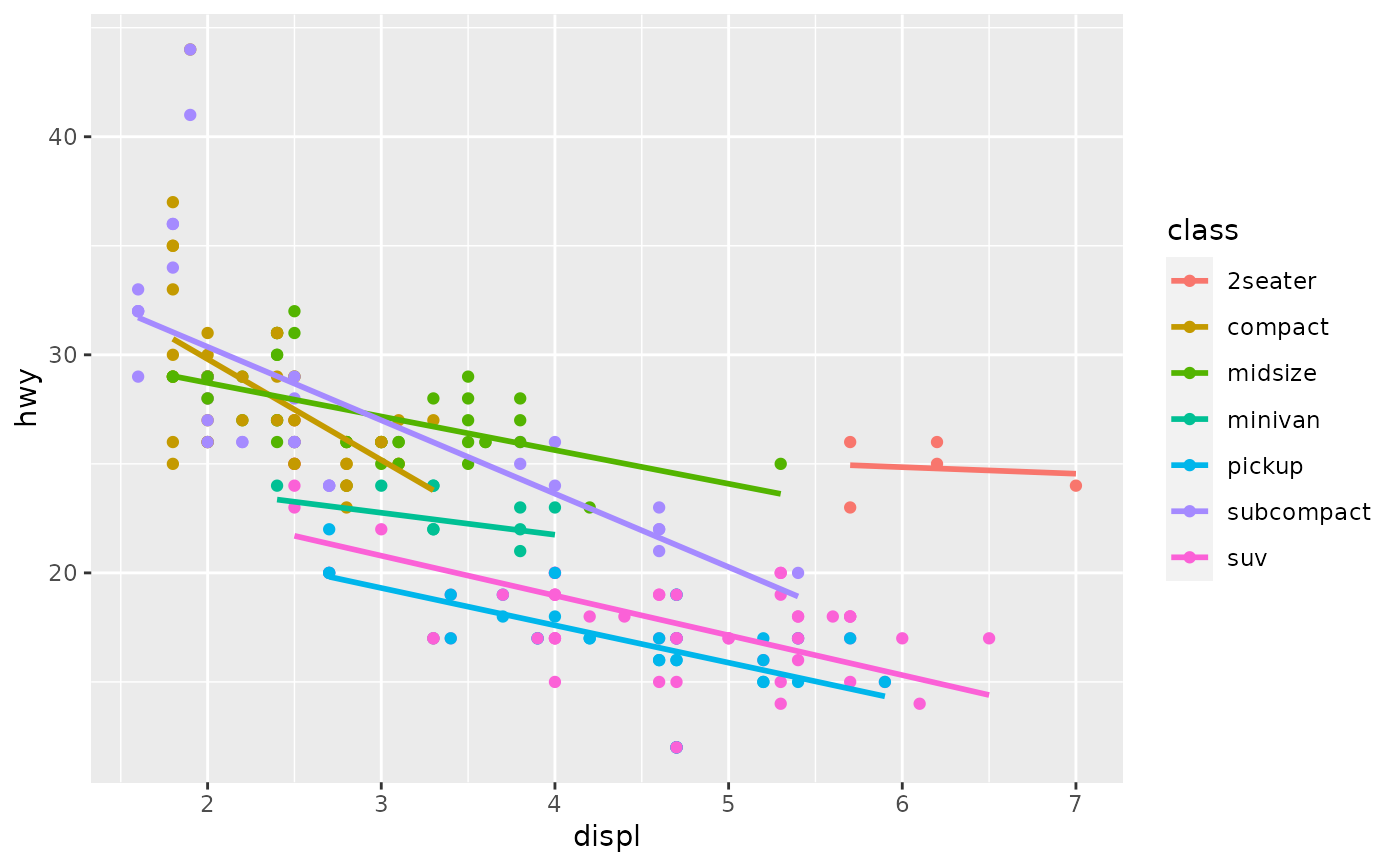
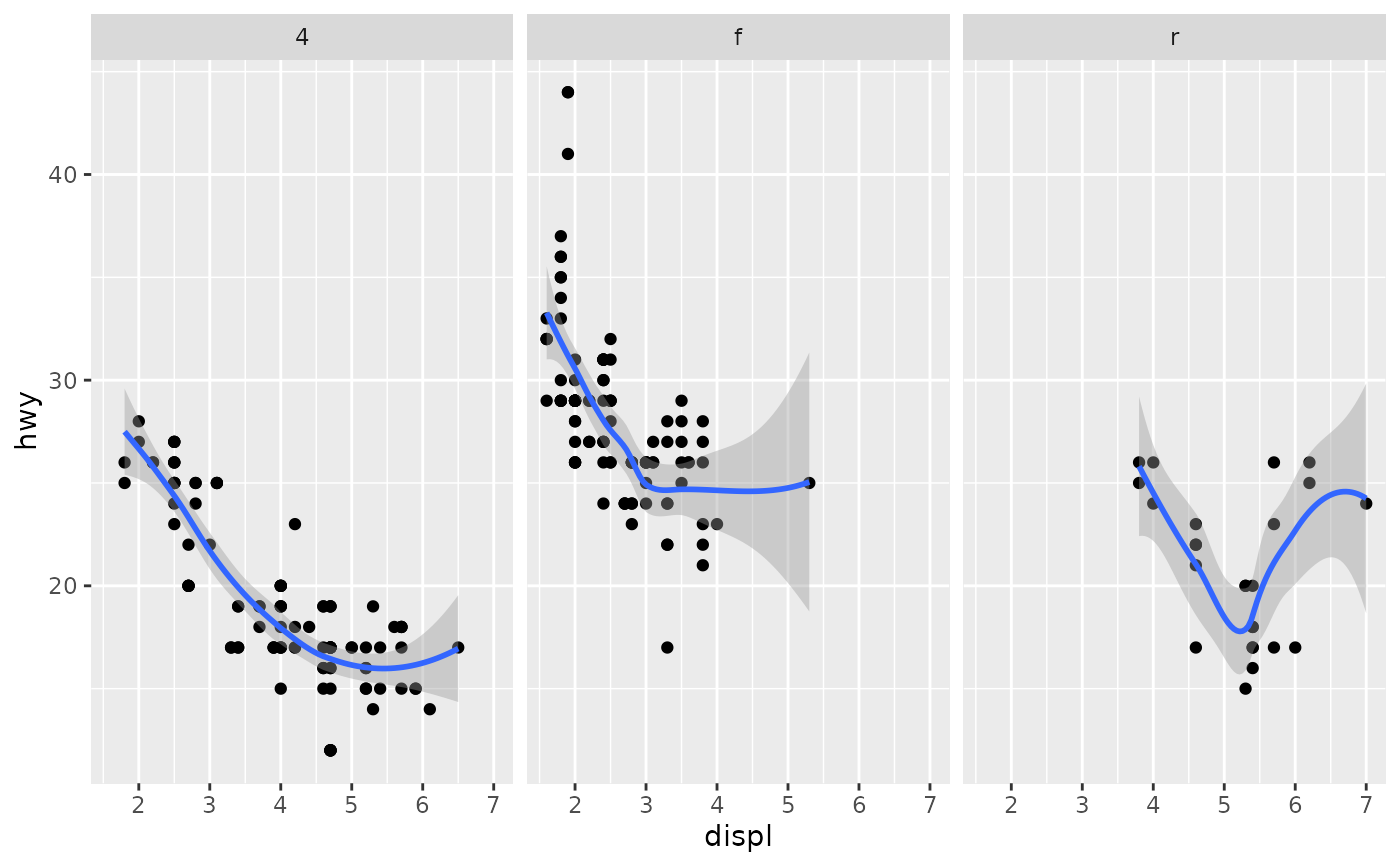
Aids the eye in seeing patterns in the presence of overplotting.
geom_smooth() and stat_smooth() are effectively aliases: they
both use the same arguments. Use stat_smooth() if you want to
display the results with a non-standard geom.
geom_smooth( mapping = NULL, data = NULL, stat = "smooth", position = "identity", ..., method = NULL, formula = NULL, se = TRUE, na.rm = FALSE, orientation = NA, show.legend = NA, inherit.aes = TRUE ) stat_smooth( mapping = NULL, data = NULL, geom = "smooth", position = "identity", ..., method = NULL, formula = NULL, se = TRUE, n = 80, span = 0.75, fullrange = FALSE, level = 0.95, method.args = list(), na.rm = FALSE, orientation = NA, show.legend = NA, inherit.aes = TRUE )
Arguments
| mapping | Set of aesthetic mappings created by |
|---|---|
| data | The data to be displayed in this layer. There are three options: If A A |
| position | Position adjustment, either as a string, or the result of a call to a position adjustment function. |
| ... | Other arguments passed on to |
| method | Smoothing method (function) to use, accepts either
For If you have fewer than 1,000 observations but want to use the same |
| formula | Formula to use in smoothing function, eg. |
| se | Display confidence interval around smooth? ( |
| na.rm | If |
| orientation | The orientation of the layer. The default ( |
| show.legend | logical. Should this layer be included in the legends?
|
| inherit.aes | If |
| geom, stat | Use to override the default connection between
|
| n | Number of points at which to evaluate smoother. |
| span | Controls the amount of smoothing for the default loess smoother.
Smaller numbers produce wigglier lines, larger numbers produce smoother
lines. Only used with loess, i.e. when |
| fullrange | Should the fit span the full range of the plot, or just the data? |
| level | Level of confidence interval to use (0.95 by default). |
| method.args | List of additional arguments passed on to the modelling
function defined by |
Details
Calculation is performed by the (currently undocumented)
predictdf() generic and its methods. For most methods the standard
error bounds are computed using the predict() method -- the
exceptions are loess(), which uses a t-based approximation, and
glm(), where the normal confidence interval is constructed on the link
scale and then back-transformed to the response scale.
Orientation
This geom treats each axis differently and, thus, can thus have two orientations. Often the orientation is easy to deduce from a combination of the given mappings and the types of positional scales in use. Thus, ggplot2 will by default try to guess which orientation the layer should have. Under rare circumstances, the orientation is ambiguous and guessing may fail. In that case the orientation can be specified directly using the orientation parameter, which can be either "x" or "y". The value gives the axis that the geom should run along, "x" being the default orientation you would expect for the geom.
Aesthetics
geom_smooth() understands the following aesthetics (required aesthetics are in bold):
xyalphacolourfillgrouplinetypesizeweightymaxymin
Learn more about setting these aesthetics in vignette("ggplot2-specs").
Computed variables
stat_smooth() provides the following variables, some of which depend on the orientation:
- y or x
predicted value
- ymin or xmin
lower pointwise confidence interval around the mean
- ymax or xmax
upper pointwise confidence interval around the mean
- se
standard error
See also
See individual modelling functions for more details:
lm() for linear smooths,
glm() for generalised linear smooths, and
loess() for local smooths.
Examples
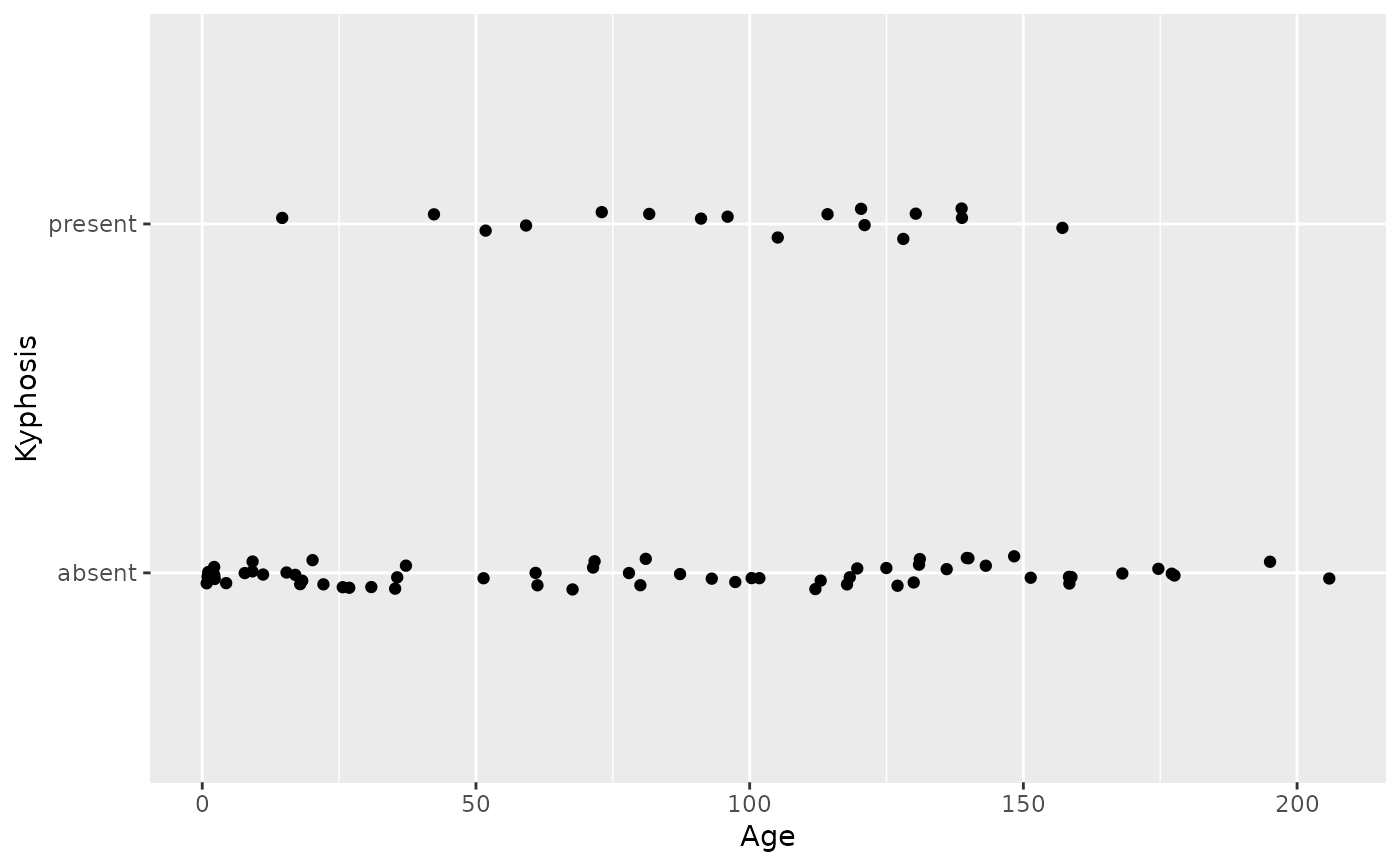
#># If you need the fitting to be done along the y-axis set the orientation ggplot(mpg, aes(displ, hwy)) + geom_point() + geom_smooth(orientation = "y")#># Use span to control the "wiggliness" of the default loess smoother. # The span is the fraction of points used to fit each local regression: # small numbers make a wigglier curve, larger numbers make a smoother curve. ggplot(mpg, aes(displ, hwy)) + geom_point() + geom_smooth(span = 0.3)#># Instead of a loess smooth, you can use any other modelling function: ggplot(mpg, aes(displ, hwy)) + geom_point() + geom_smooth(method = lm, se = FALSE)#>ggplot(mpg, aes(displ, hwy)) + geom_point() + geom_smooth(method = lm, formula = y ~ splines::bs(x, 3), se = FALSE)# Smooths are automatically fit to each group (defined by categorical # aesthetics or the group aesthetic) and for each facet. ggplot(mpg, aes(displ, hwy, colour = class)) + geom_point() + geom_smooth(se = FALSE, method = lm)#>#># \donttest{ binomial_smooth <- function(...) { geom_smooth(method = "glm", method.args = list(family = "binomial"), ...) } # To fit a logistic regression, you need to coerce the values to # a numeric vector lying between 0 and 1. ggplot(rpart::kyphosis, aes(Age, Kyphosis)) + geom_jitter(height = 0.05) + binomial_smooth()#>#> Warning: Computation failed in `stat_smooth()`: #> y values must be 0 <= y <= 1ggplot(rpart::kyphosis, aes(Age, as.numeric(Kyphosis) - 1)) + geom_jitter(height = 0.05) + binomial_smooth()#>ggplot(rpart::kyphosis, aes(Age, as.numeric(Kyphosis) - 1)) + geom_jitter(height = 0.05) + binomial_smooth(formula = y ~ splines::ns(x, 2))# But in this case, it's probably better to fit the model yourself # so you can exercise more control and see whether or not it's a good model. # }