This function also allows you to assign different designs to different groups or individuals in a population.
design(x, deslist = list(), descol = character(0), ...)
Arguments
| x | model object |
|---|---|
| deslist | a list of |
| descol | the |
| ... | not used |
Details
This setup requires the use of an idata_set, with individual-level
data passed in one ID per row. For each ID, specify a
grouping variable in idata (descol). For each unique value
of the grouping variable, make one tgrid object and pass them
in order as ... or form them into a list and pass as deslist.
You must assign the idata_set before assigning the designs in the
command chain (see the example below).
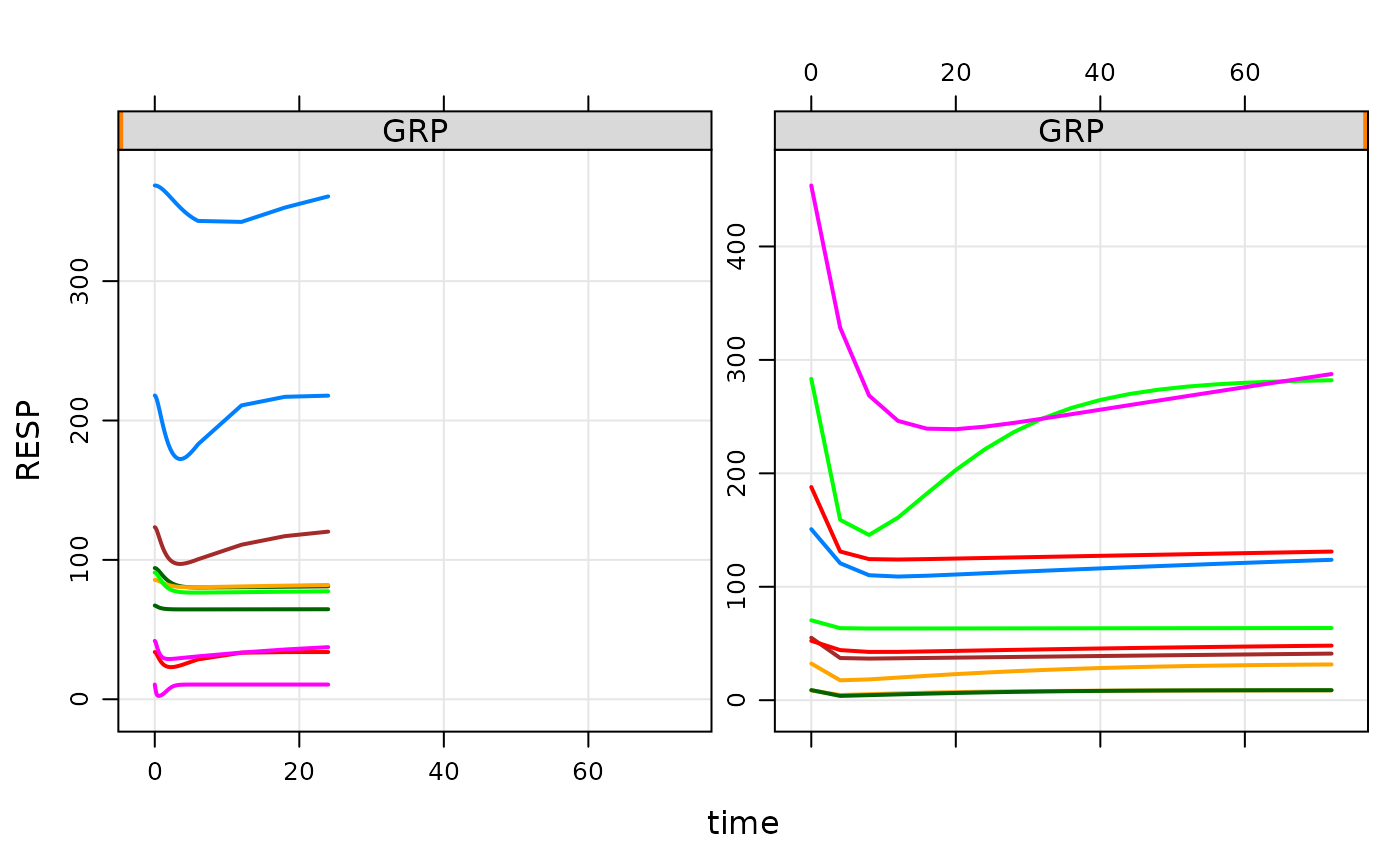
Examples
peak <- tgrid(0,6,0.1) sparse <- tgrid(0,24,6) des1 <- c(peak,sparse) des2 <- tgrid(0,72,4) data <- expand.ev(ID = 1:10, amt=c(100,300)) data$GRP <- data$amt/100 idata <- data[,c("ID", "amt")] mod <- mrgsolve::house() mod %>% omat(dmat(1,1,1,1)) %>% carry_out(GRP) %>% idata_set(idata) %>% design(list(des1, des2),"amt") %>% data_set(data) %>% mrgsim %>% plot(RESP~time|GRP)